Help and Documents
Introduction
Circular RNAs (circRNAs) are stable, single-stranded RNA molecules produced through back splicing. Lack of free ends making them resistant to degradation by exonucleases. Due to the high stability and tissue-specific expression, circRNAs are promising targets for clinical research. They have various functions such as binding to the proteins and regulation of gene expression. Their role as competing endogenous RNA (ceRNA) is extensively studied. CircPac prepares a user-friendly environment to examine and visualize circRNAs.
CircPac Key Features
CircPac provides three different types of analytical tools for users, including circRNA ID Retrieval, Examination, and Data Visualization.
circRNA ID Retrieval
In this section, there is the BLAST tool. It's capable of taking single or multiple circRNA sequences and returns the corresponding circBase-IDs. It also converts the result into an Excel file which can be delivered to the user.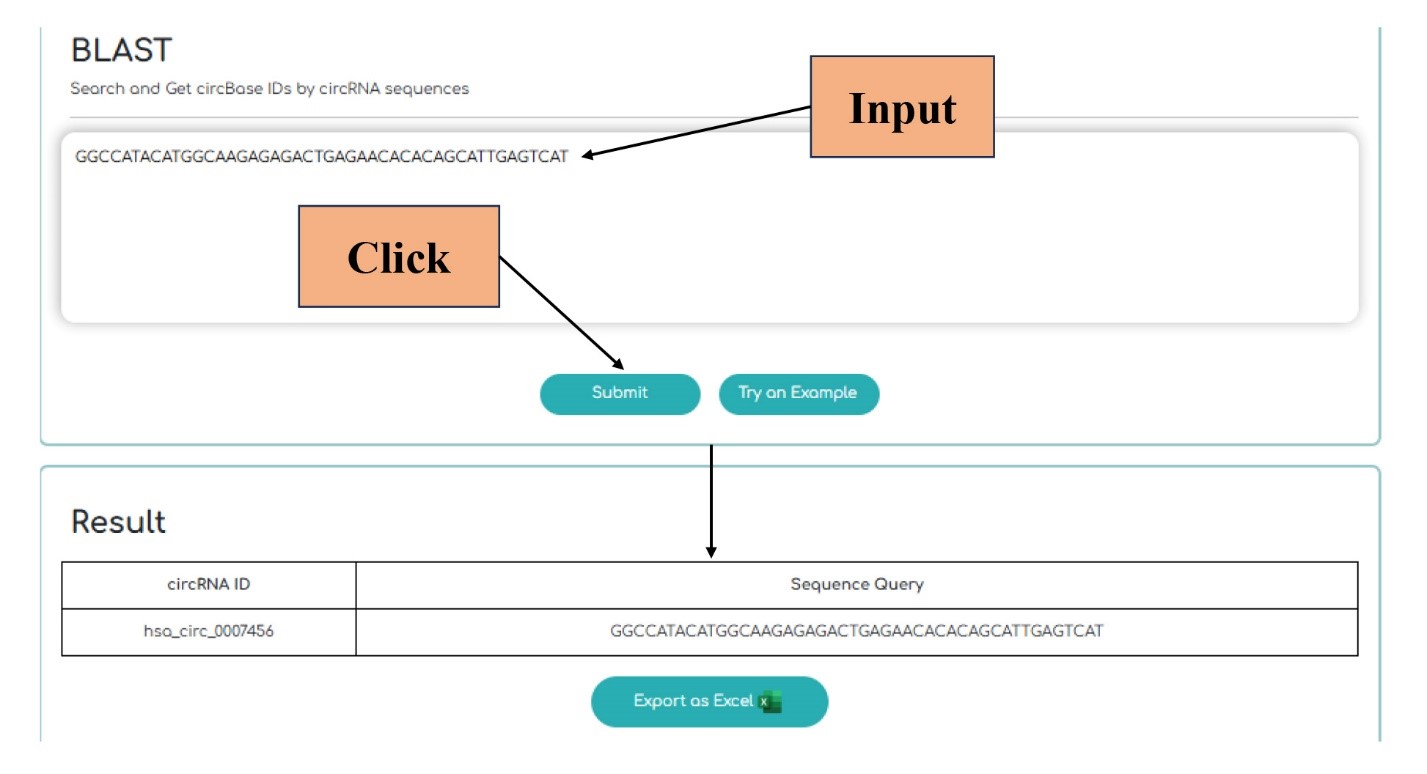
Examination
By inputting the circBase-IDs acquired in the previous step, the user can get information using three tools: "circRNA Information," "circRNA Expression" and "miRNA interactions". “circRNA Information” tool provides basic information about circRNAs, including genomic positions, strands, genomic length, spliced length, and gene symbol. "circRNA Expression" and “miRNA interactions” reveal expression changes of circRNAs in diseases and miRNAs interacting with circRNAs, respectively. The results can be presented in Excel file format.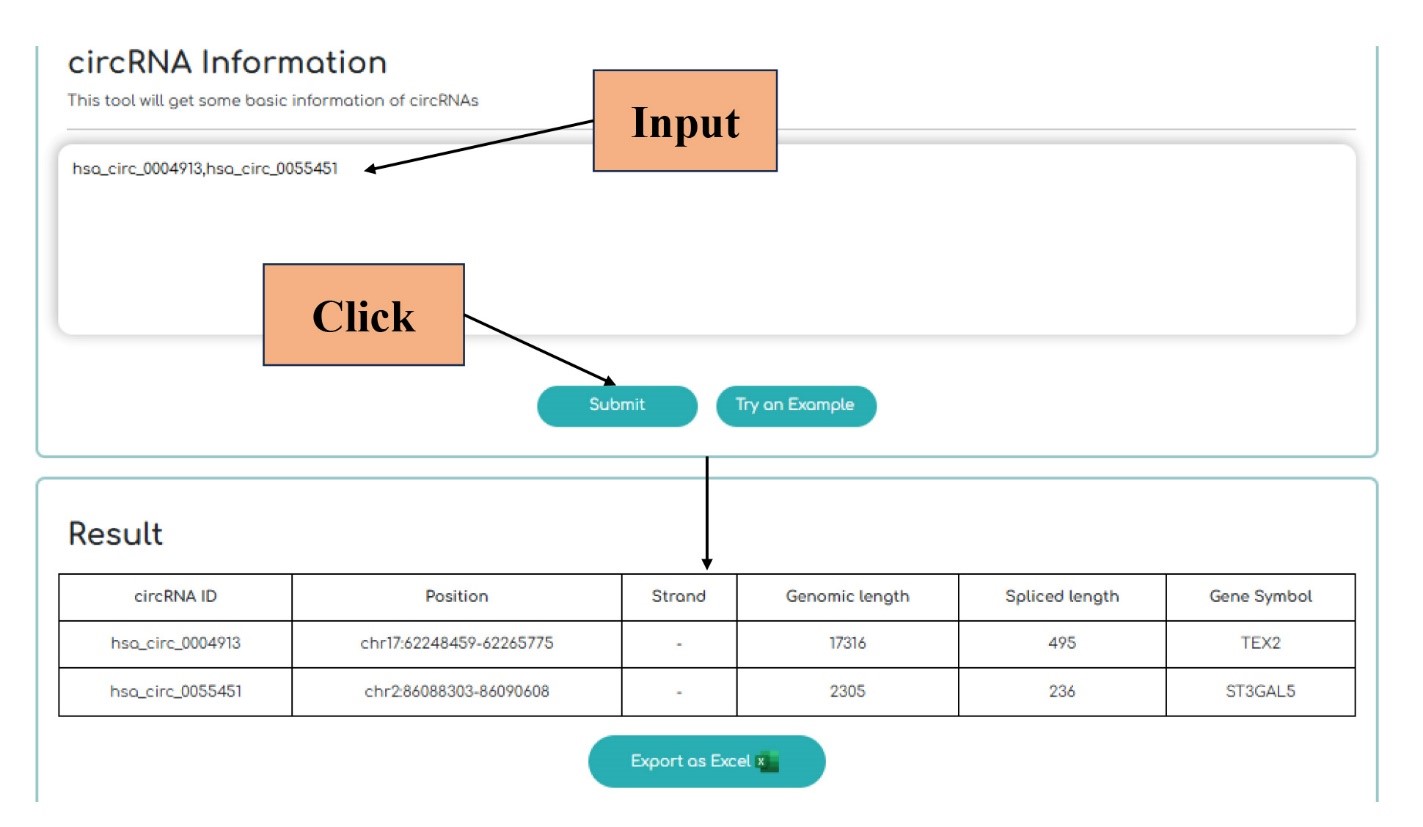
Data Visualization
This category of tools is for visualization of the investigated data, which include the following two modules: “circRNA Expression Plot” and “miRNA Interactions Plot” tools. “circRNA Expression Plot” tool provides heatmap diagram indicating circRNA expression changes in diseases. “miRNA Interactions Plot” tool shows miRNA-circRNA interactions by the circular chord diagram style.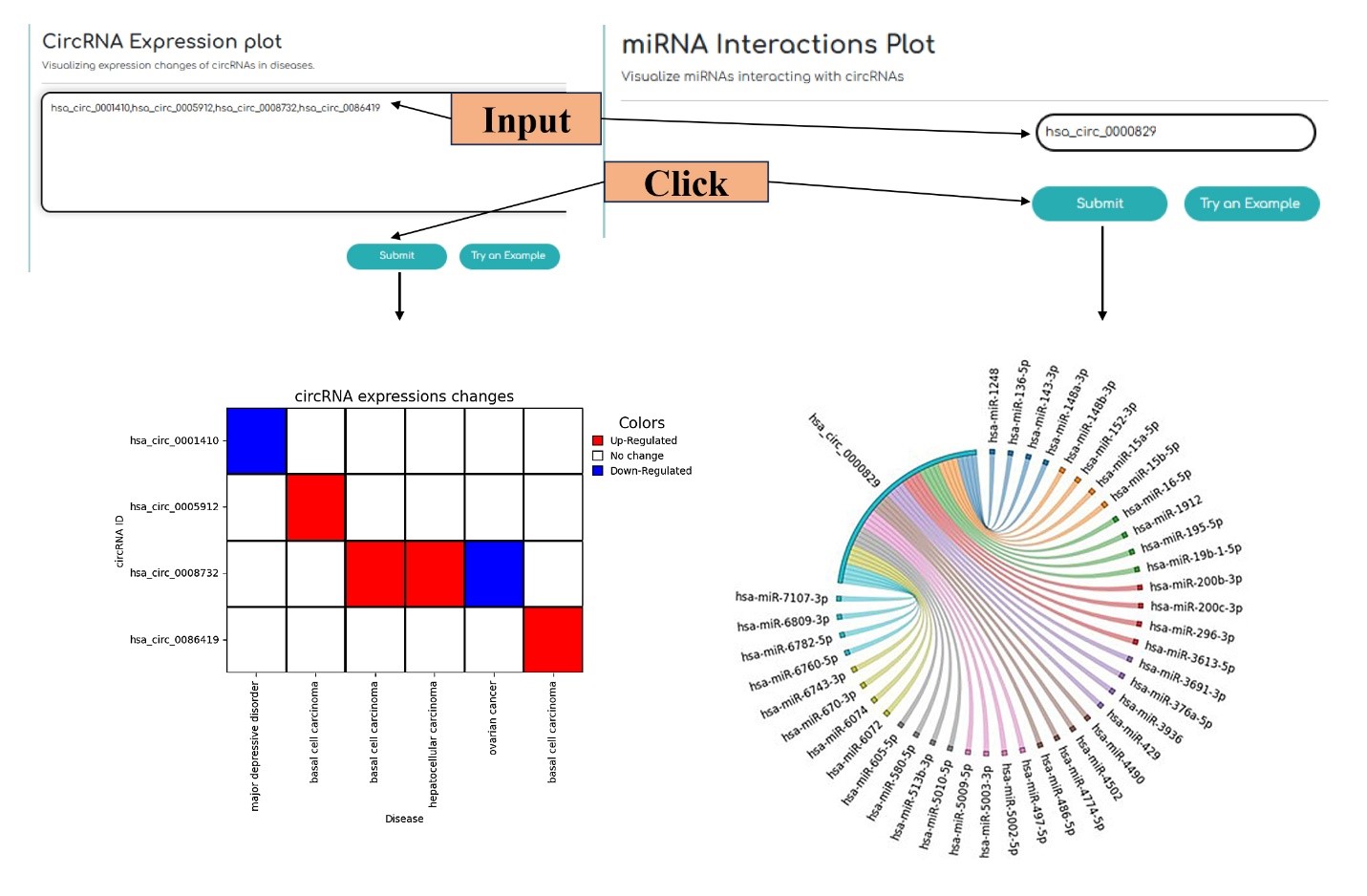
Data Collection and Implementation
The data of CircPac originates from biological databases including circBase (http://www.circbase.org/), circBank (www.circbank.cn), circRNADisease (http://cgga.org.cn/circRNADisease/), and CircFunBase (http://bis.zju.edu.cn/CircFunBase). CircPac utilized the downloadable files from circBank, circRNADisease, circBase databases, and for CircFunBase, it accessed its data through the respective Application Programming Interface (API). The analysis and comparisons were conducted with the data obtained without any intervention. The BLAST tool conducts search in the CircFunBase database one by one and provides the corresponding circBase-IDs for each query. To acquire basic information about circRNAs, it utilizes circBase. It also retrieves circRNA expression changes data from circRNADisease and extracts details on miRNA-circRNA interactions from circBank.
circPac is developed by the CircPac team at Isfahan university of medical sciences and Isfahan University, Isfahan, Iran
First version: February 2024
Last Update: January 2025